Plot Objects
Plot-objects.RmdOverview
The lionfish package offers multiple different types of display elements. These have to be provided to the interactive_tour function that launches the GUI. Each plot object is a list containing a type and an object. The type specifies, which kind of display should be generated. The object provides additional information required to construct the display. The plot objects then have to be stored in a list that can be given to the interactive_tour function. The currently supported displays are:
- 1d tour
- 2d tour
- scatterplot
- histogram
- mosaic plot
- heatmap
- categorical cluster interface
Examples for all displays can be found below.
Setup
# Load required packages
library(tourr)
library(lionfish)
if (requireNamespace("flexclust")) {library(flexclust)}
#> Loading required namespace: flexclust
# Initialize python backend
if (check_venv()){
init_env(env_name = "r-lionfish", virtual_env = "virtual_env")
} else if (check_conda_env()){
init_env(env_name = "r-lionfish", virtual_env = "anaconda")
}Dataset 1 - Flea data
# Load dataset
data("flea")
# Prepare objects for later us
clusters_flea <- as.numeric(flea$species)
flea_subspecies <- unique(flea$species)
# Standardize data and calculate half_range
flea <- apply(flea[,1:6], 2, function(x) (x-mean(x))/sd(x))
feature_names_flea <- colnames(flea)
half_range_flea <- max(sqrt(rowSums(flea^2)))Dataset 2 - Winter activities data
if (requireNamespace("flexclust")) {
# Load dataset and set seed
data("winterActiv")
set.seed(42)
# Perform kmeans clustering
clusters_winter <- stepcclust(winterActiv, k=6, nrep=20)
clusters_winter <- clusters_winter@cluster
# Get the names of our features
feature_names_winter <- colnames(winterActiv)
}
#> 6 : * * * * * * * * * * * * * * * * * * * *Currently supported display types
One dimensional tour
To display one dimensional tours, they first have to be generated and saved using the save_history function of the tourr package. For more information on the available types of tours please visit tour path construction.
Example plot object
guided_tour_flea_1d <- save_history(flea,
tour_path=guided_tour(holes(),1))
#> Target: 1.530, 28.9% better
#> Target: 1.607, 5.0% better
#> Target: 1.629, 1.4% better
#> Target: 1.638, 0.6% better
#> Target: 1.653, 0.9% better
#> Target: 1.656, 0.1% better
#> No better bases found after 25 tries. Giving up.
#> Final projection:
#> 0.438
#> -0.281
#> -0.247
#> -0.532
#> 0.338
#> -0.520
obj_flea_1d_tour <- list(type="1d_tour", # type of display
obj=guided_tour_flea_1d) # 1d tour history Interactivity
- Changing frames of tour: The frames of the current tour can be changed by pressing the left arrow-key for the last frame and the right arrow-key for the next frame. The current frame is displayed in the frame on the left.
- Projection manipulation: the projection can be manipulated via drag and drop using the right mouse button. (Note for Mac users using trackpads: two fingers have to be used to right-click).
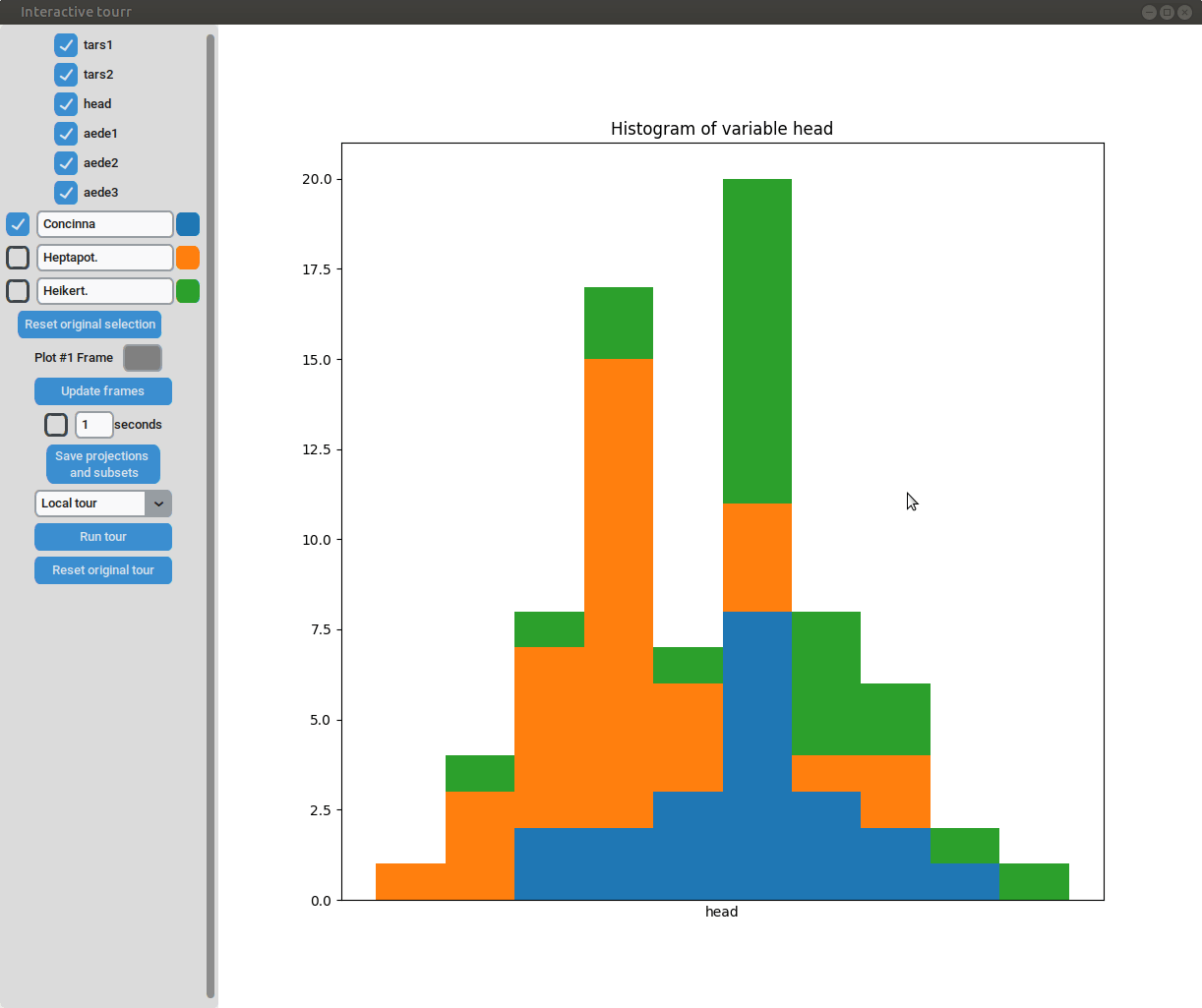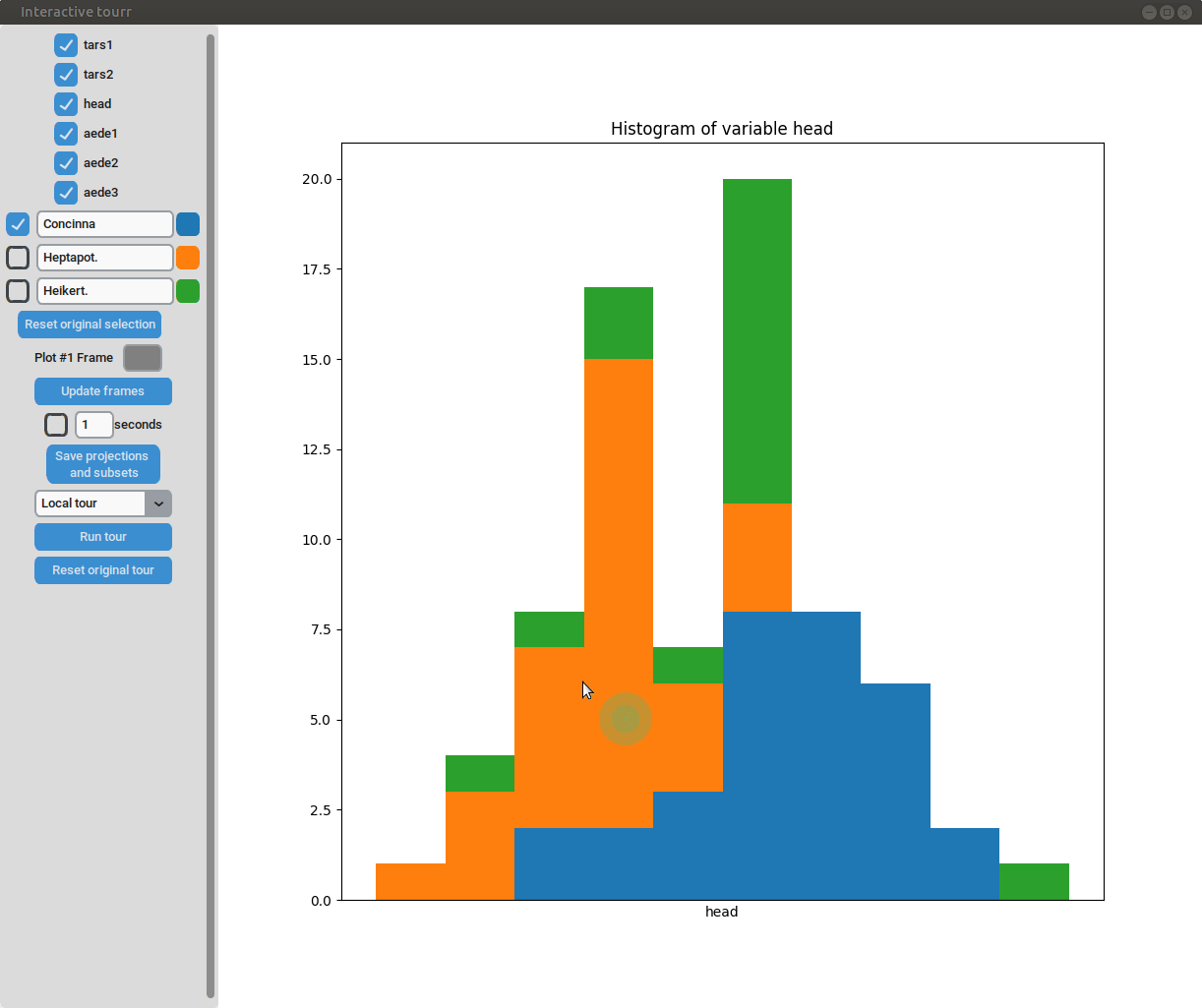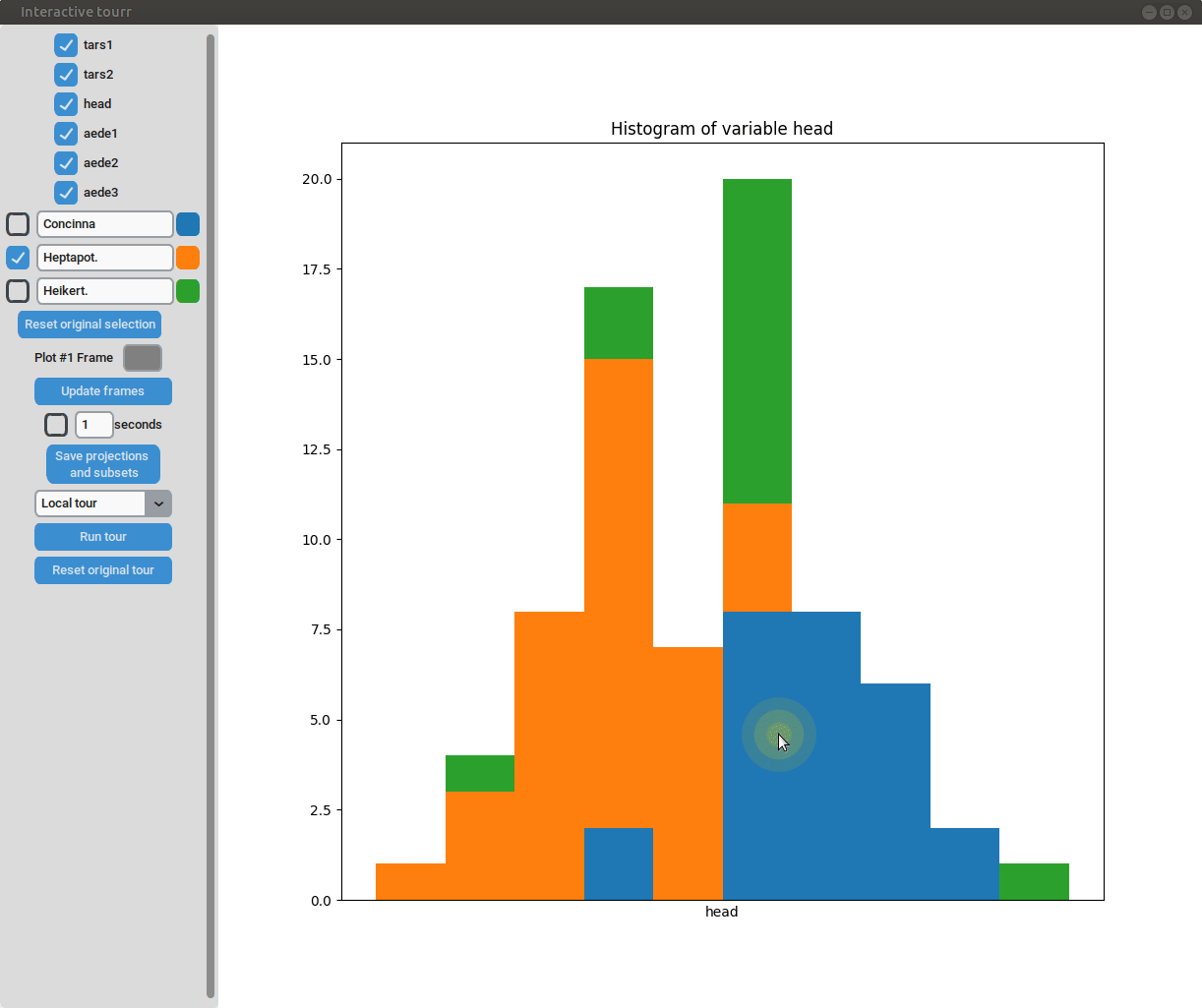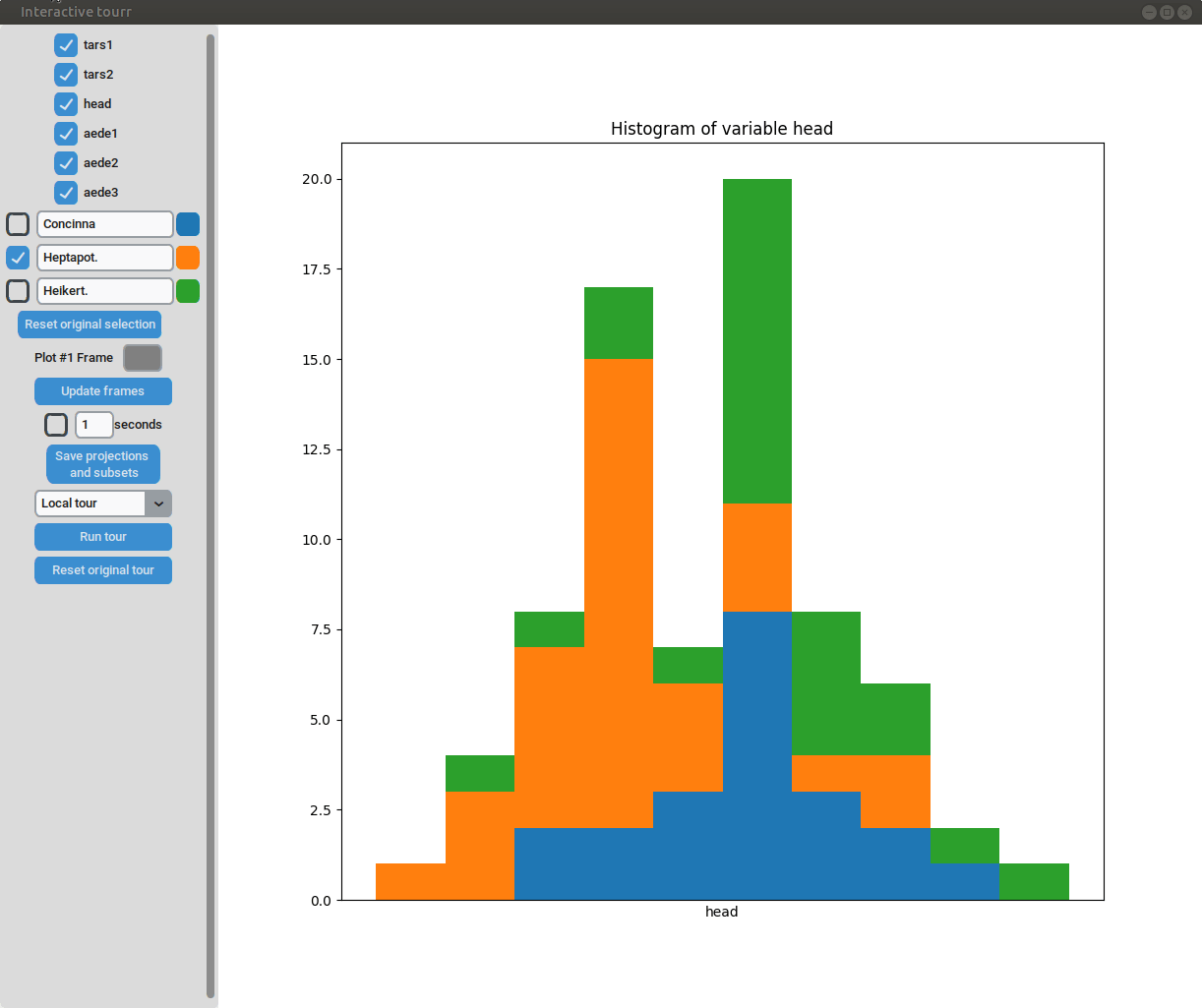
- Subselection: The datapoins within each subset can be changed by clicking on one of the bars of the histogram. The datapoints within the bar are then moved to the subset currently selected on the left.
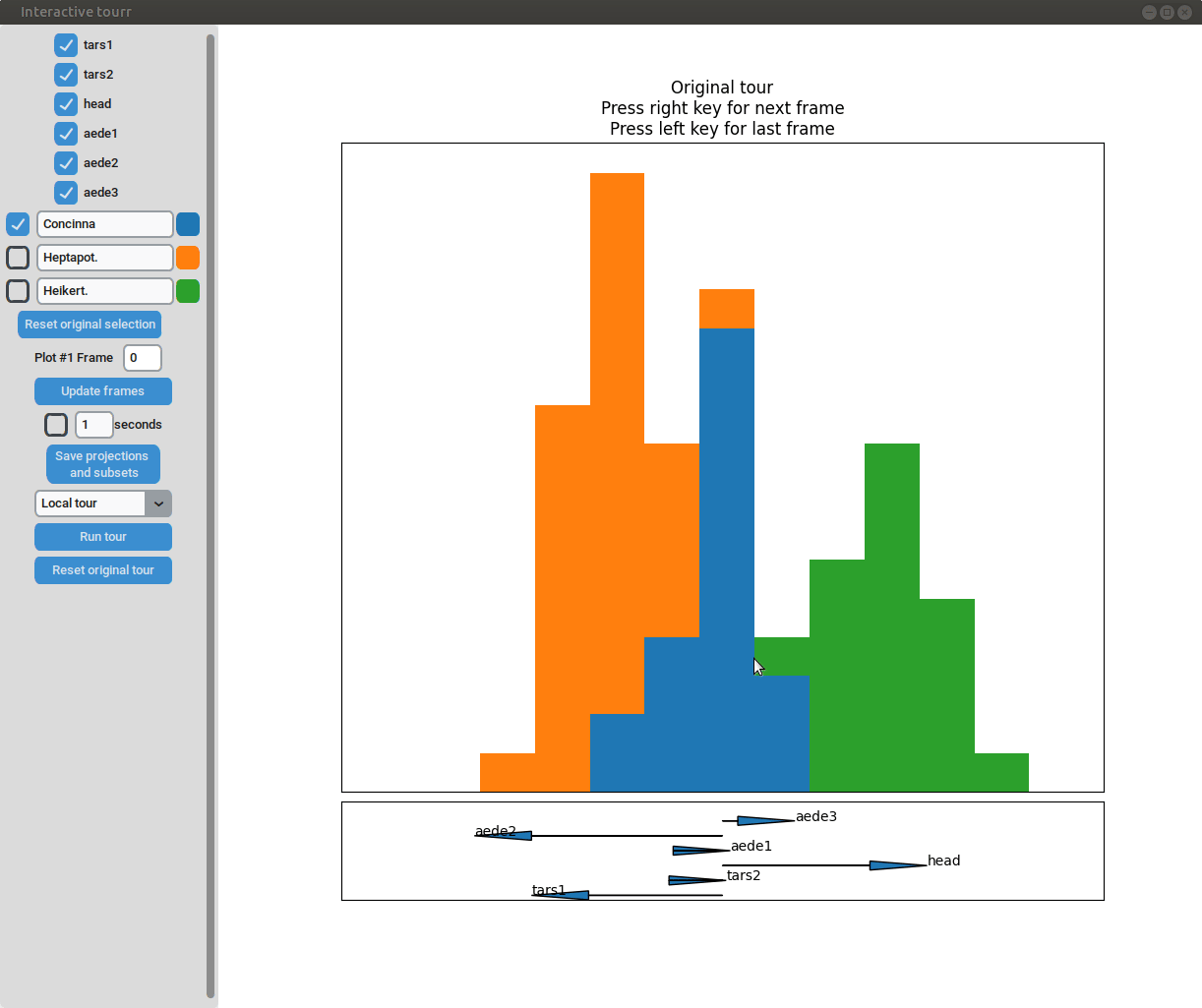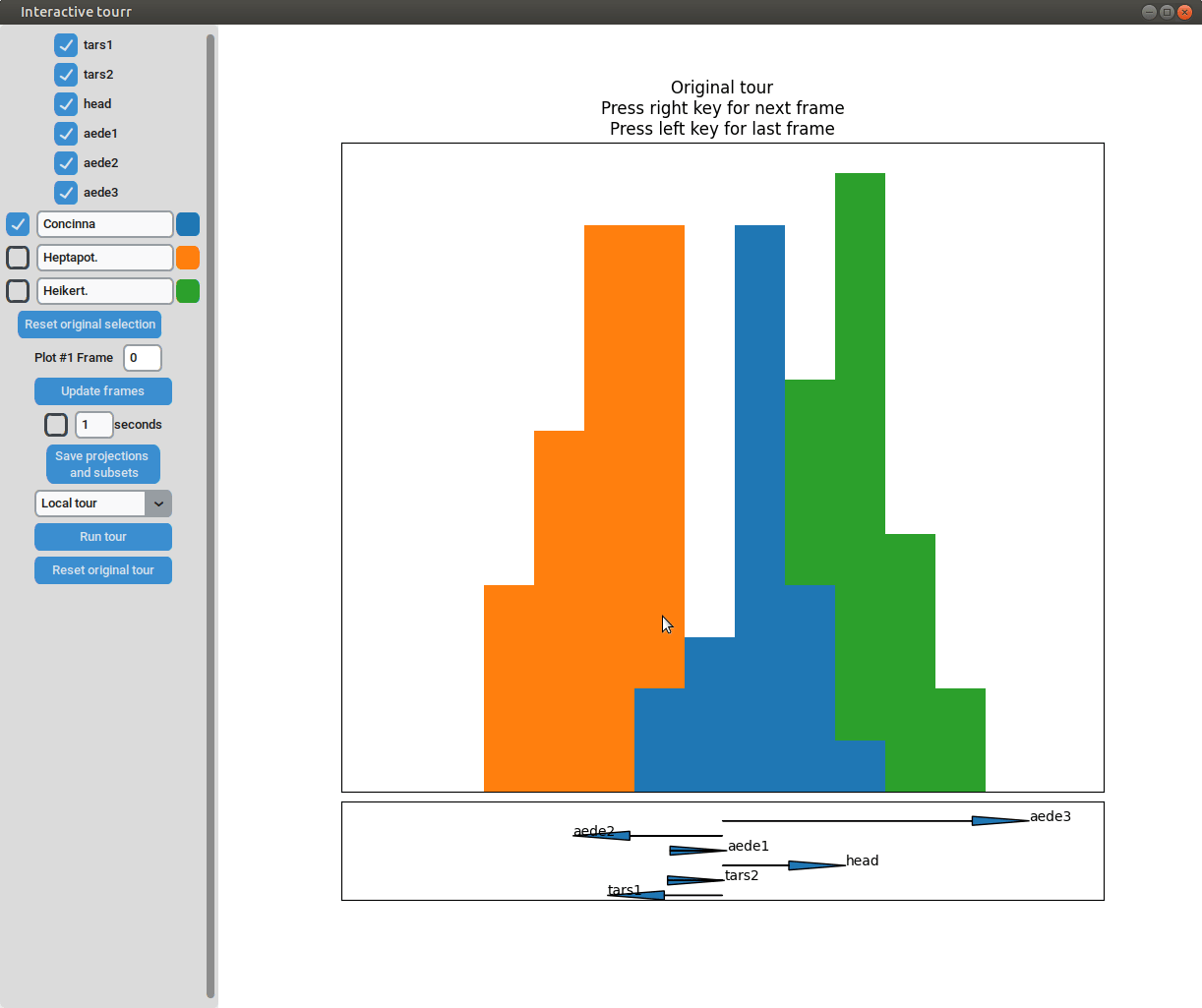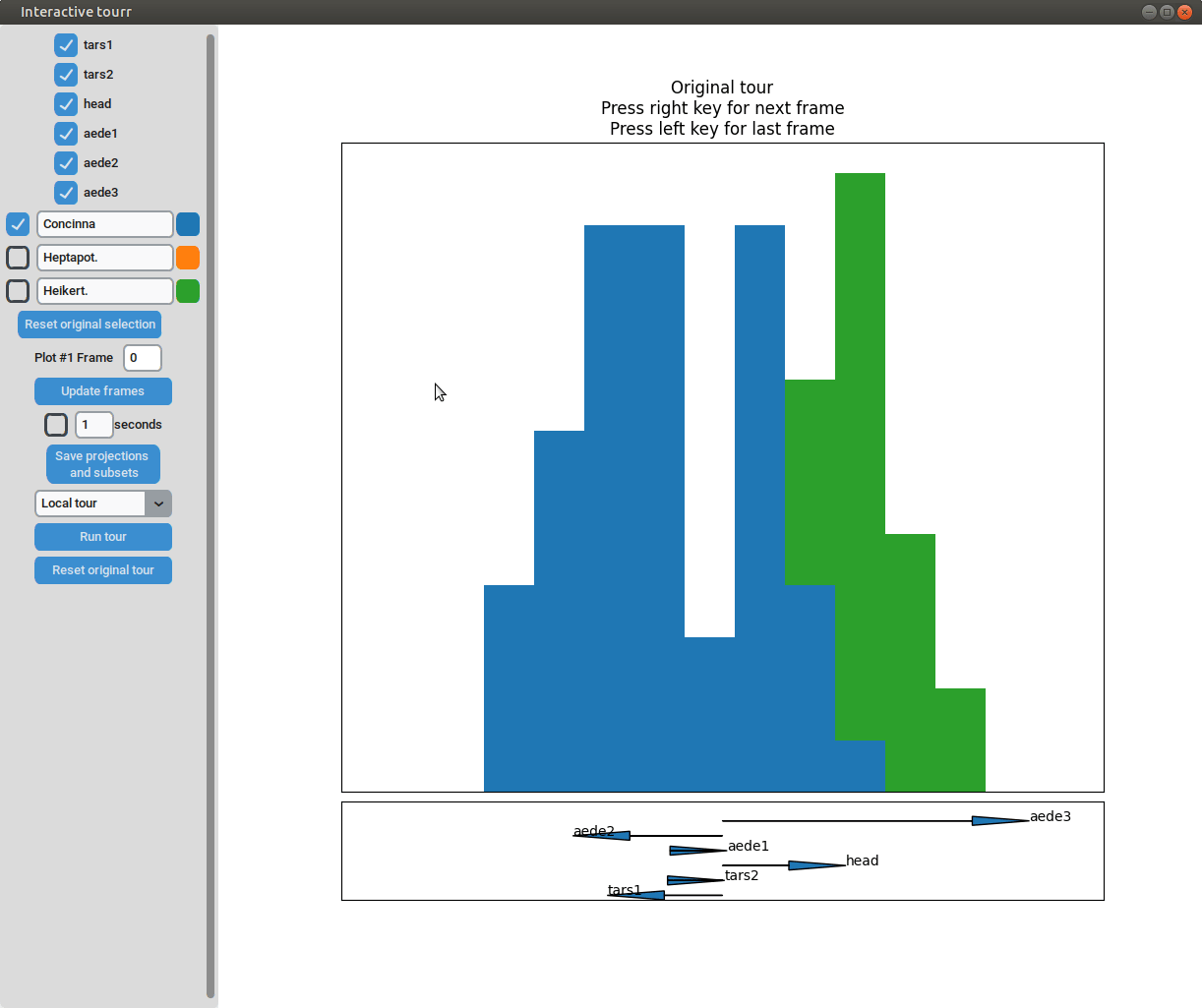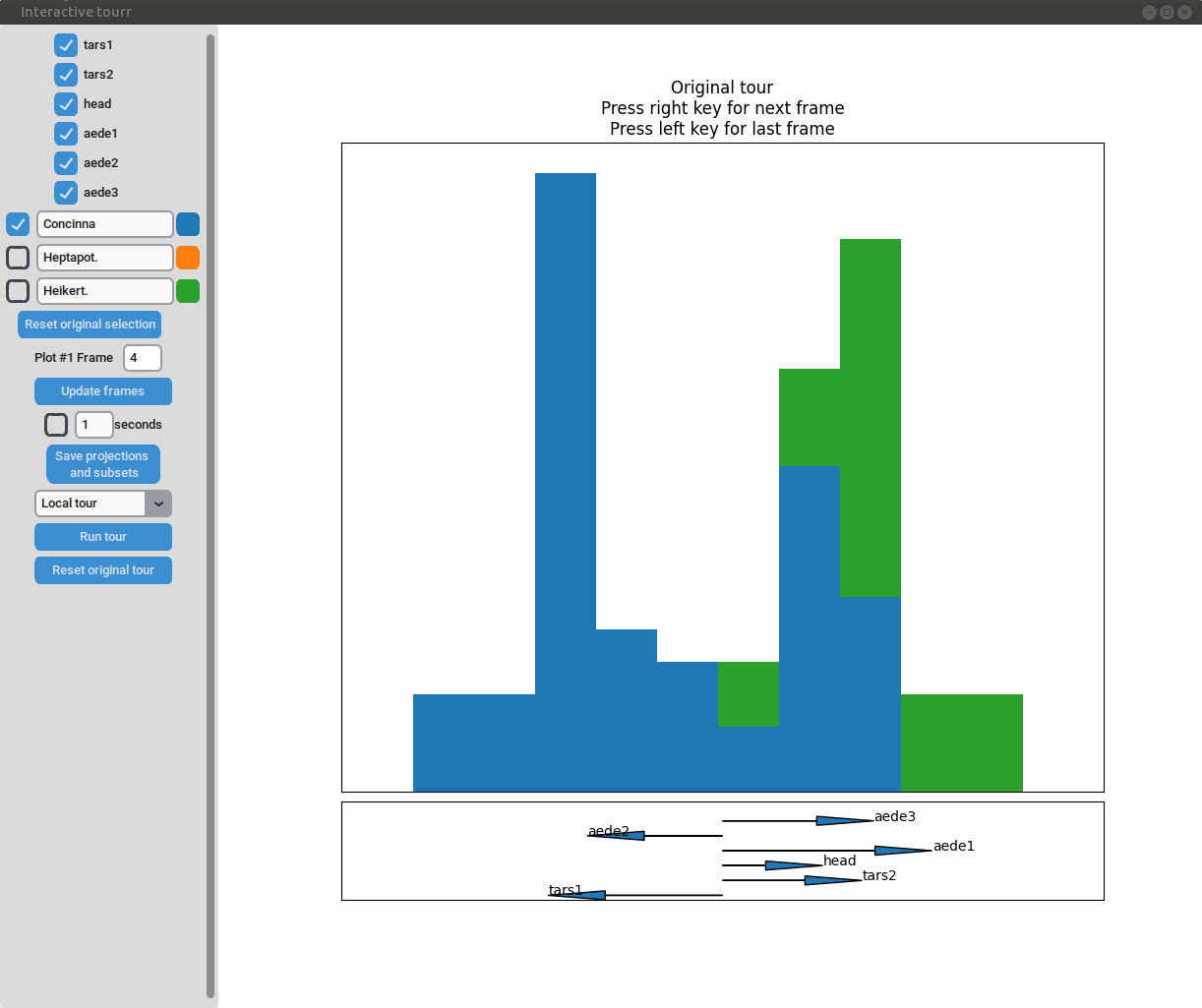
Two dimensional tour
To display two dimensional tours, they first have to be generated and saved using the save_history function of the tourr package. For more information on the available types of tours please visit tour path construction.
Example plot object
grand_tour_flea_2d <- save_history(flea,
tour_path = grand_tour(d=2))
obj_flea_2d_tour <- list(type="2d_tour", # type of display
obj=grand_tour_flea_2d) # 2d tour historyInteractivity
- Changing frames of tour: The frames of the current tour can be changed by pressing the left arrow-key for the last frame and the right arrow-key for the next frame. The current frame is displayed in the frame on the left.
- Projection manipulation: the projection can be manipulated via drag and drop using the right mouse button. (Note for Mac users using trackpads: two fingers have to be used to right-click).
- Subselection: The datapoins within each subset can be changed by pressing the left mouse-button and drawing a lasso around the datapoints to be selected. The datapoints within the lasso are then moved to the subset currently selected on the left.
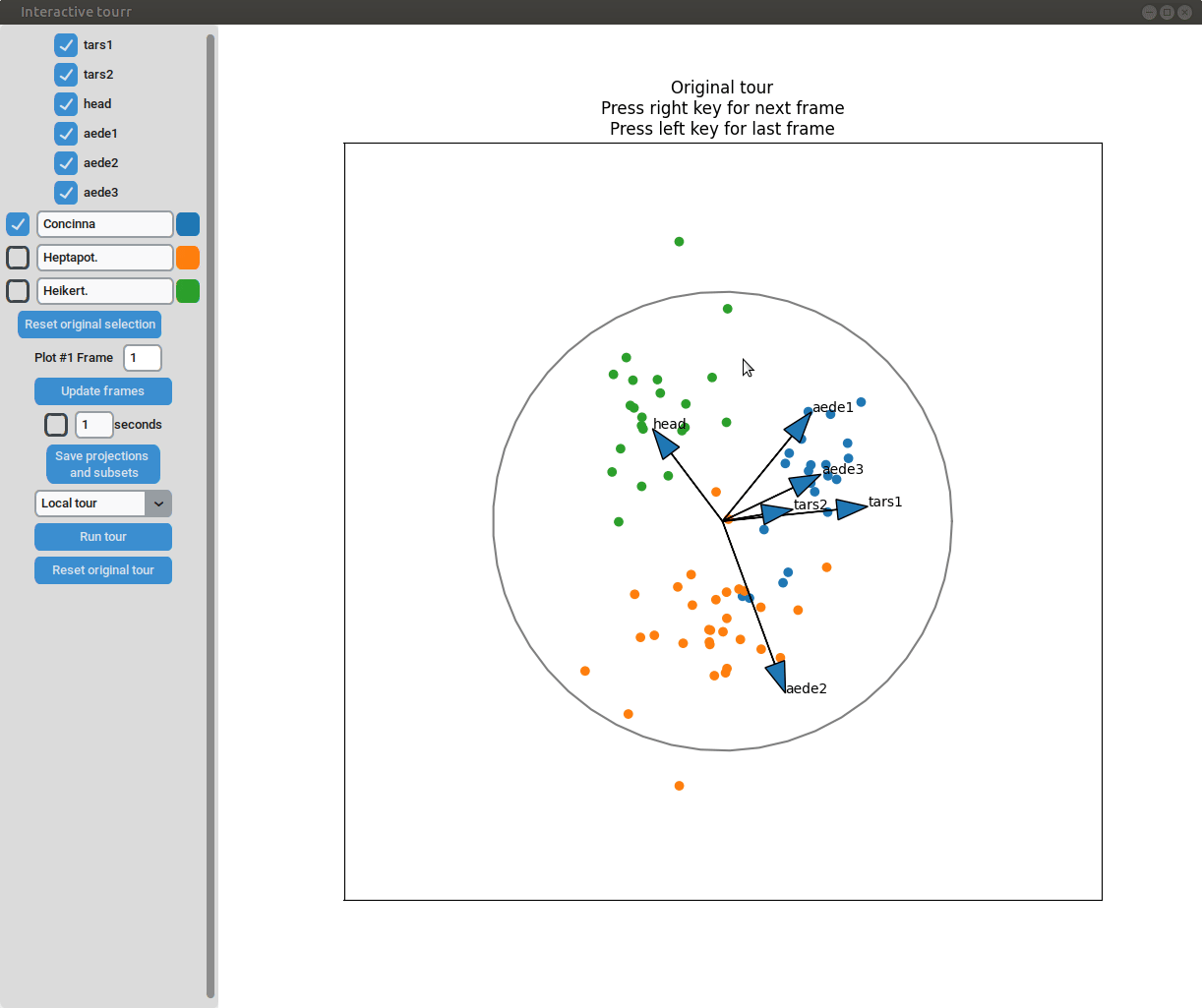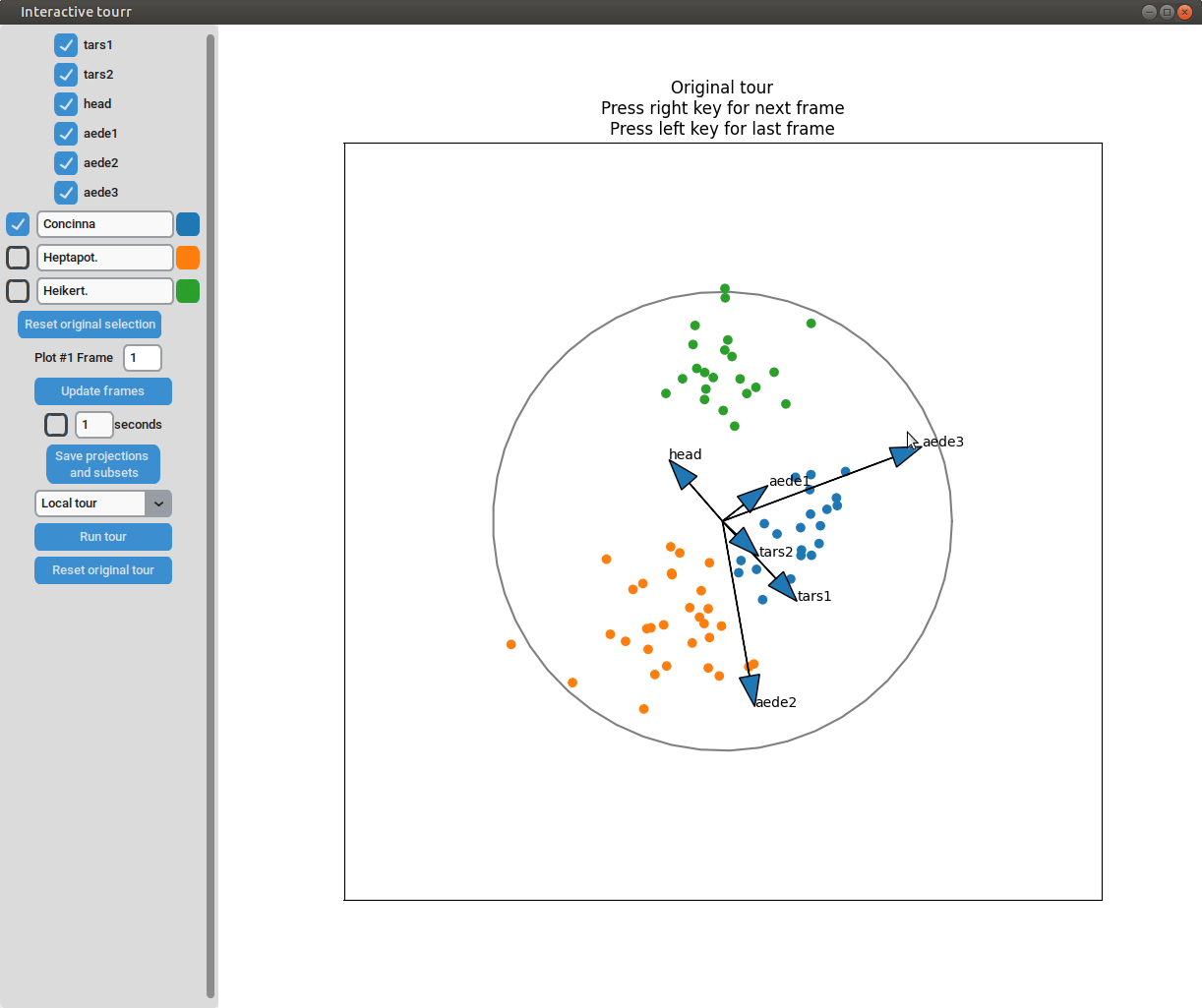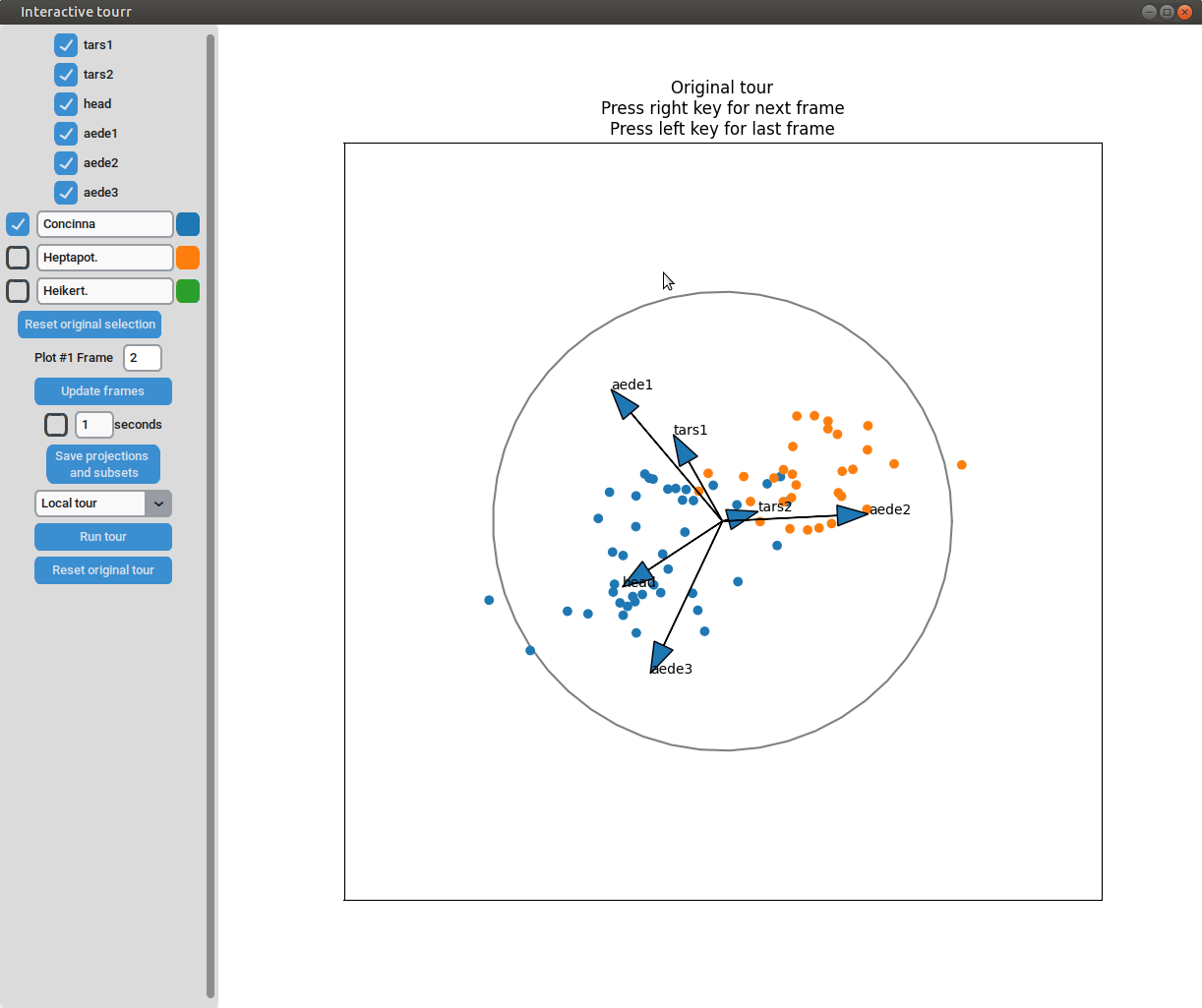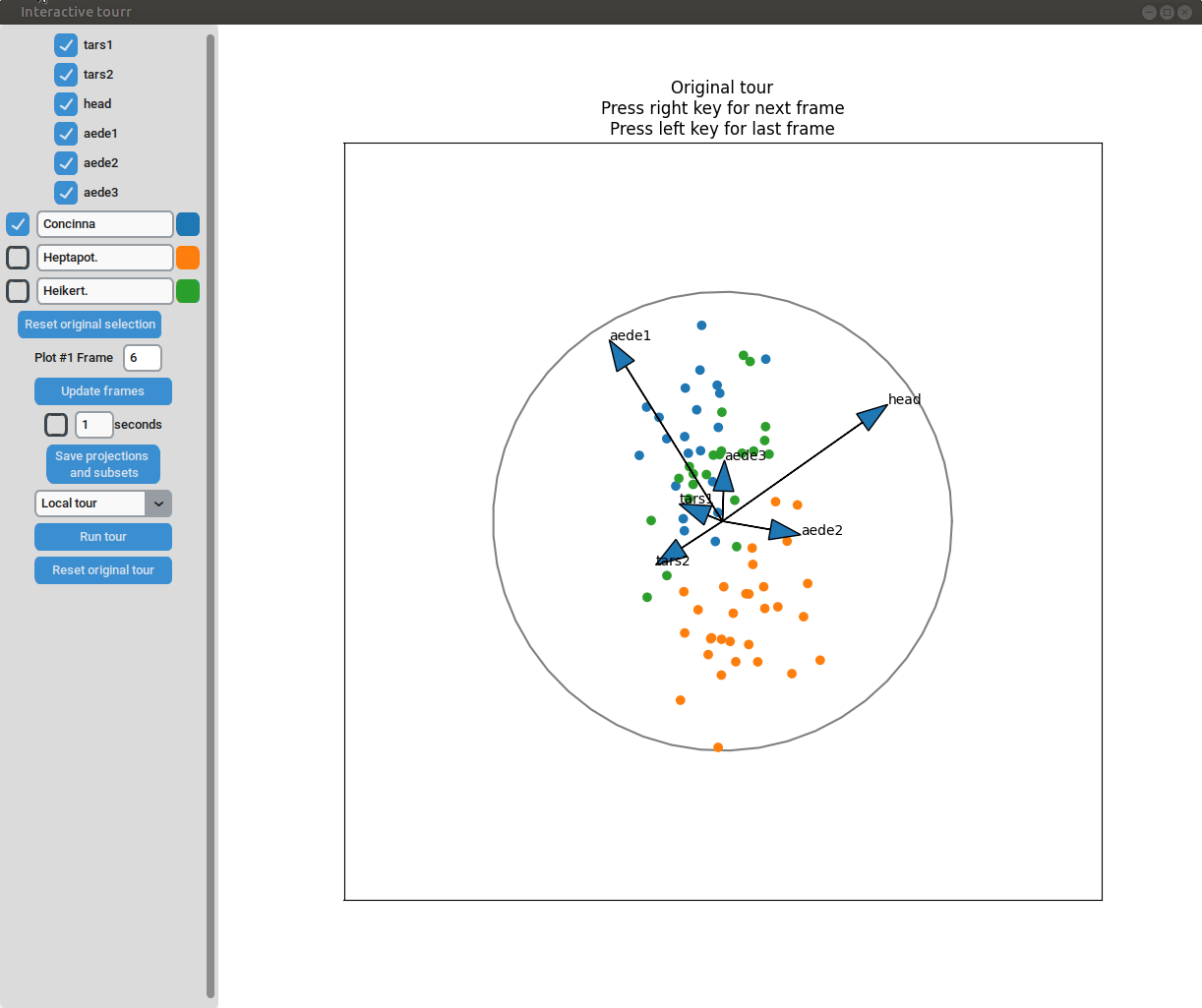
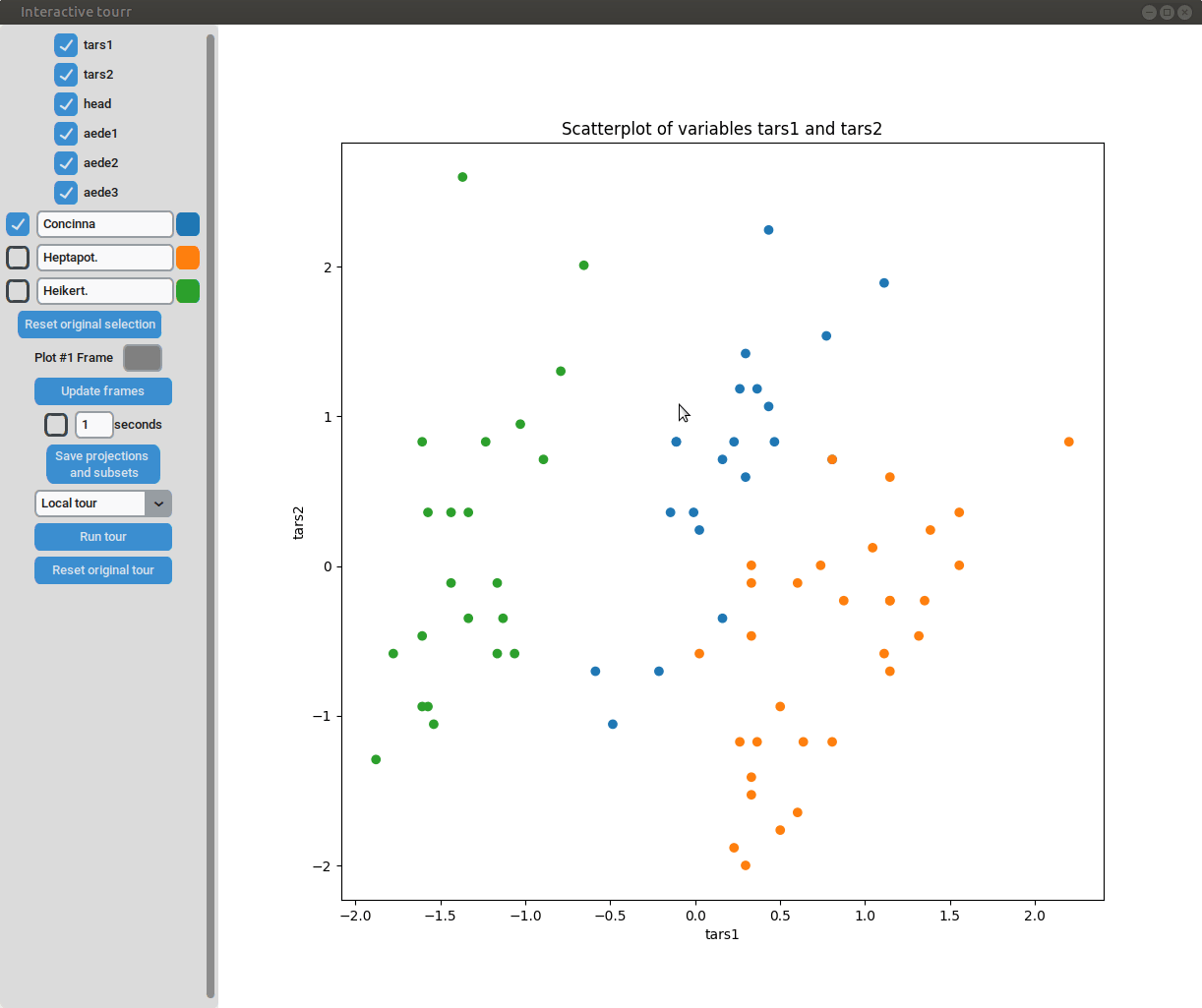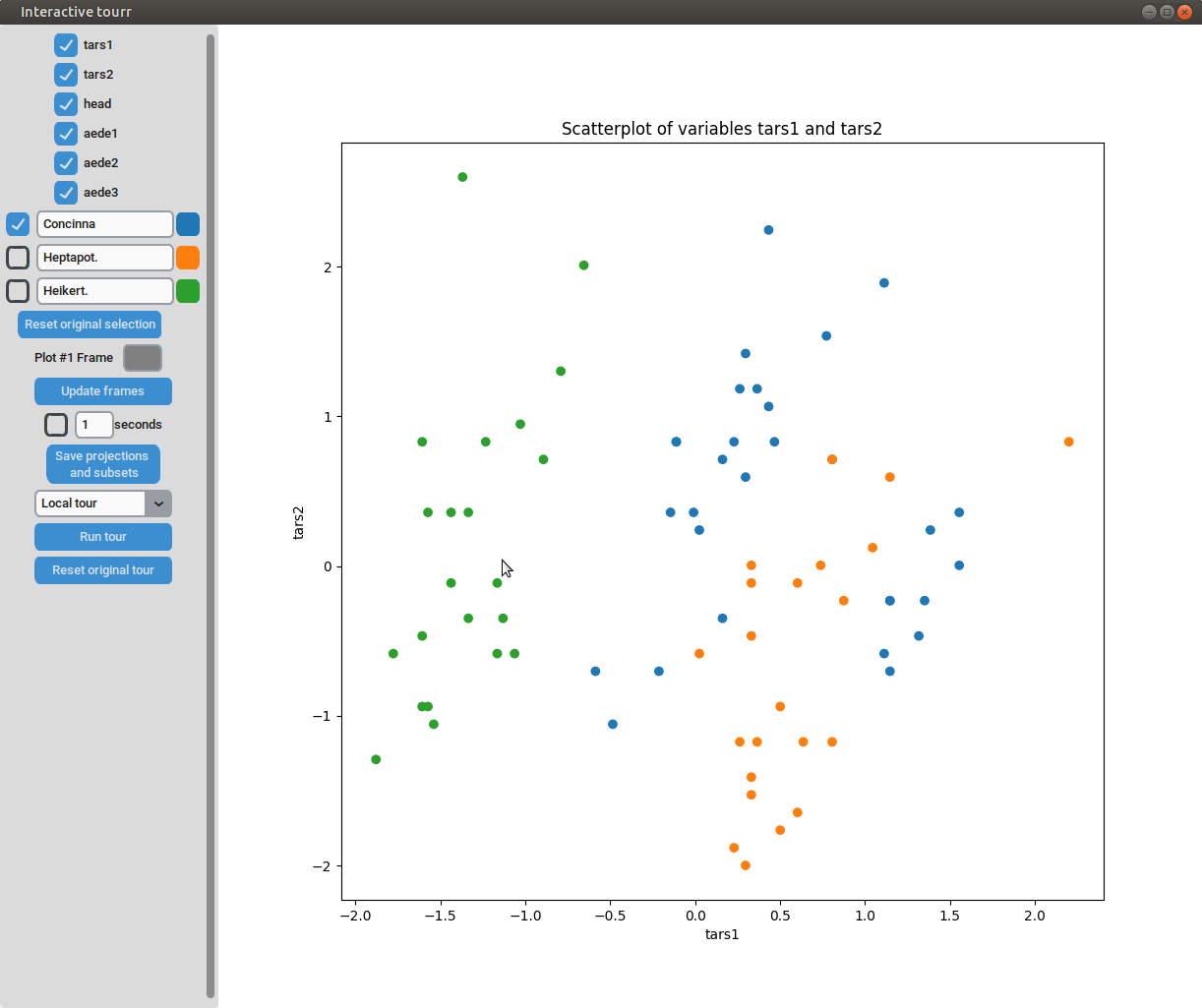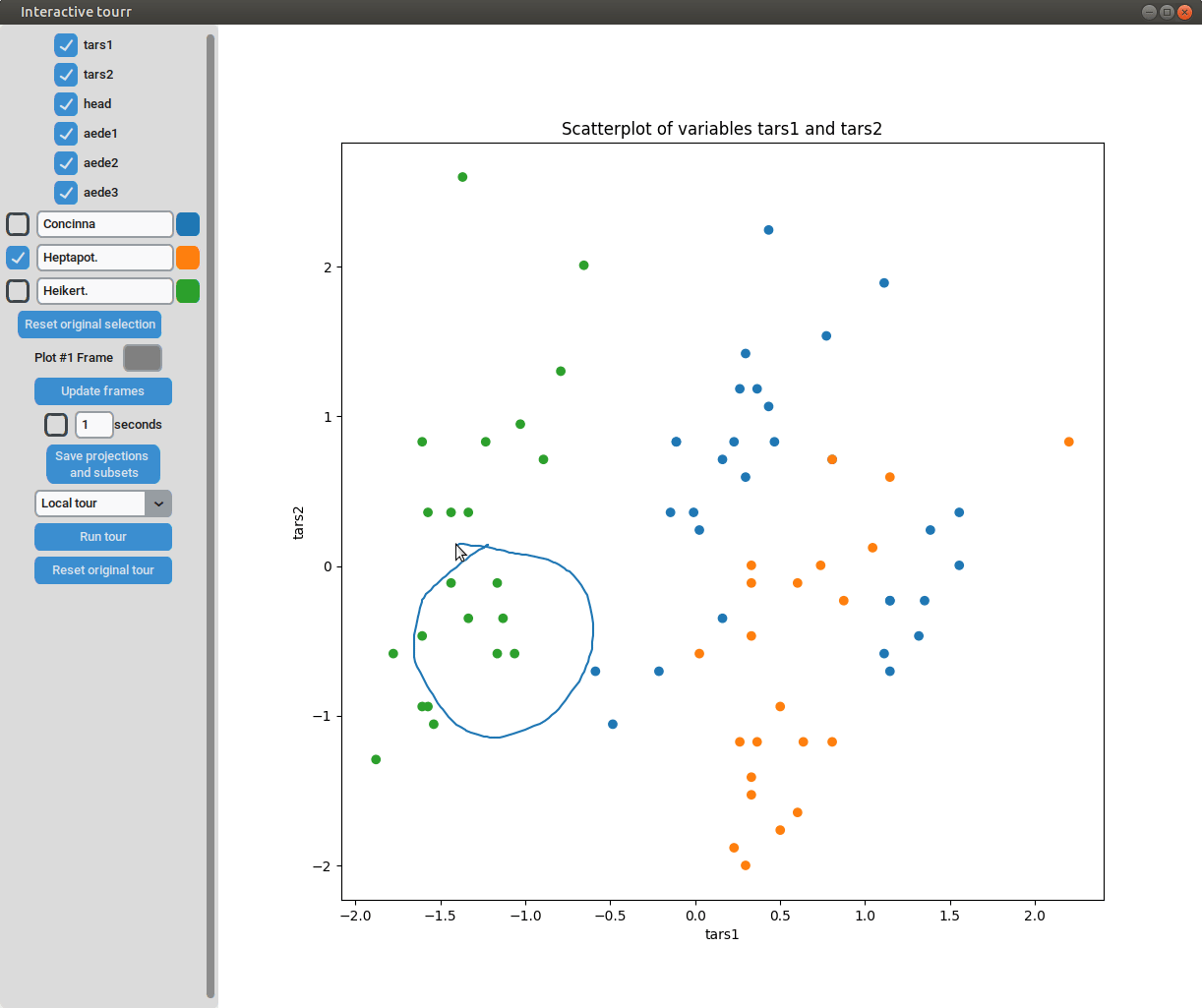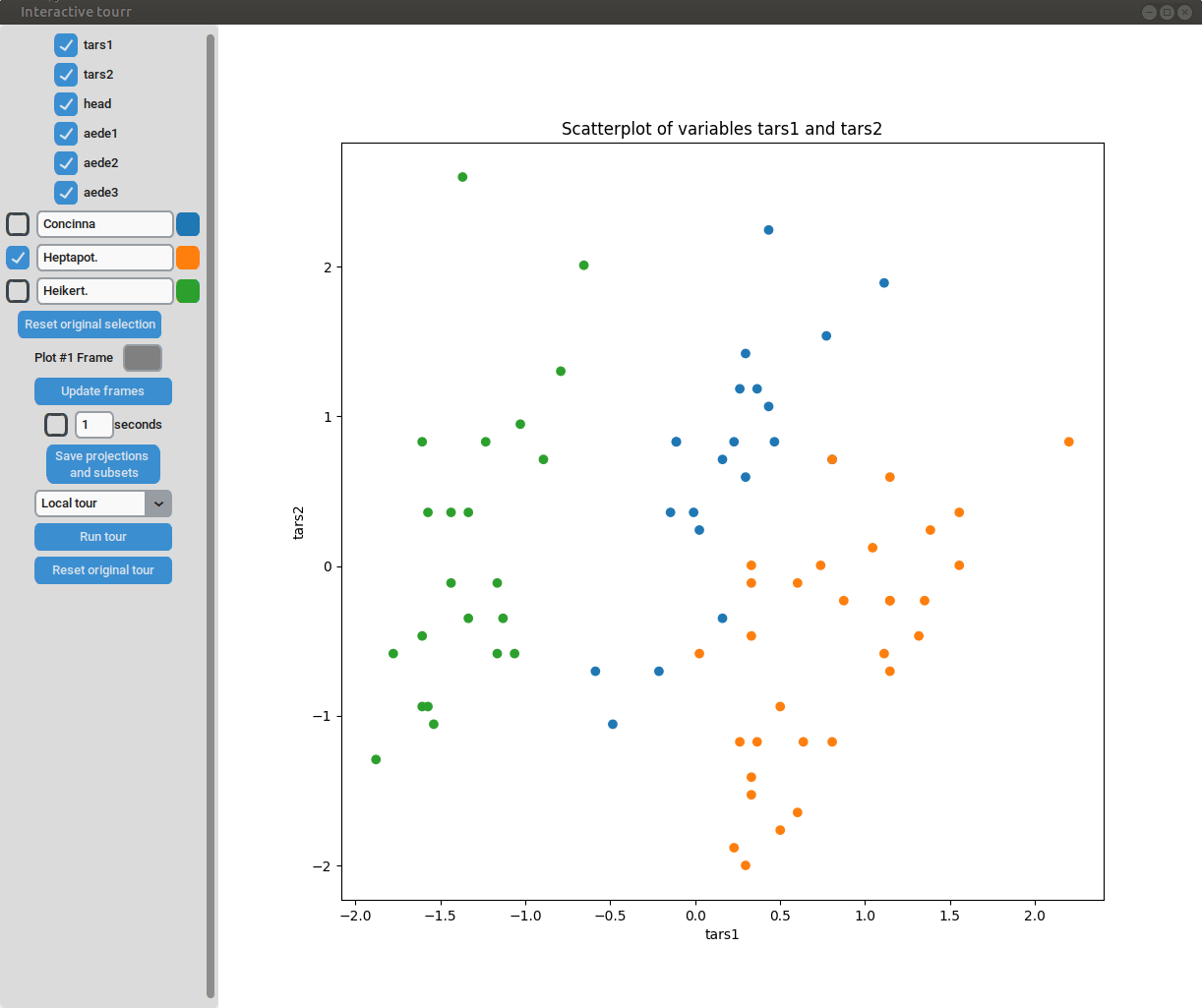
Scatterplot
To display a scatter plot, the features to be displayed on the x and y axis have to be provided in form of a two dimensional vector.
Histogram
To display a histogram, the feature to be displayed on the x axis has to be provided in form of a two dimensional vector.
Example plot object
obj_flea_histogram <- list(type="hist", # type of display
obj="head") # x axis of histogramMosaic
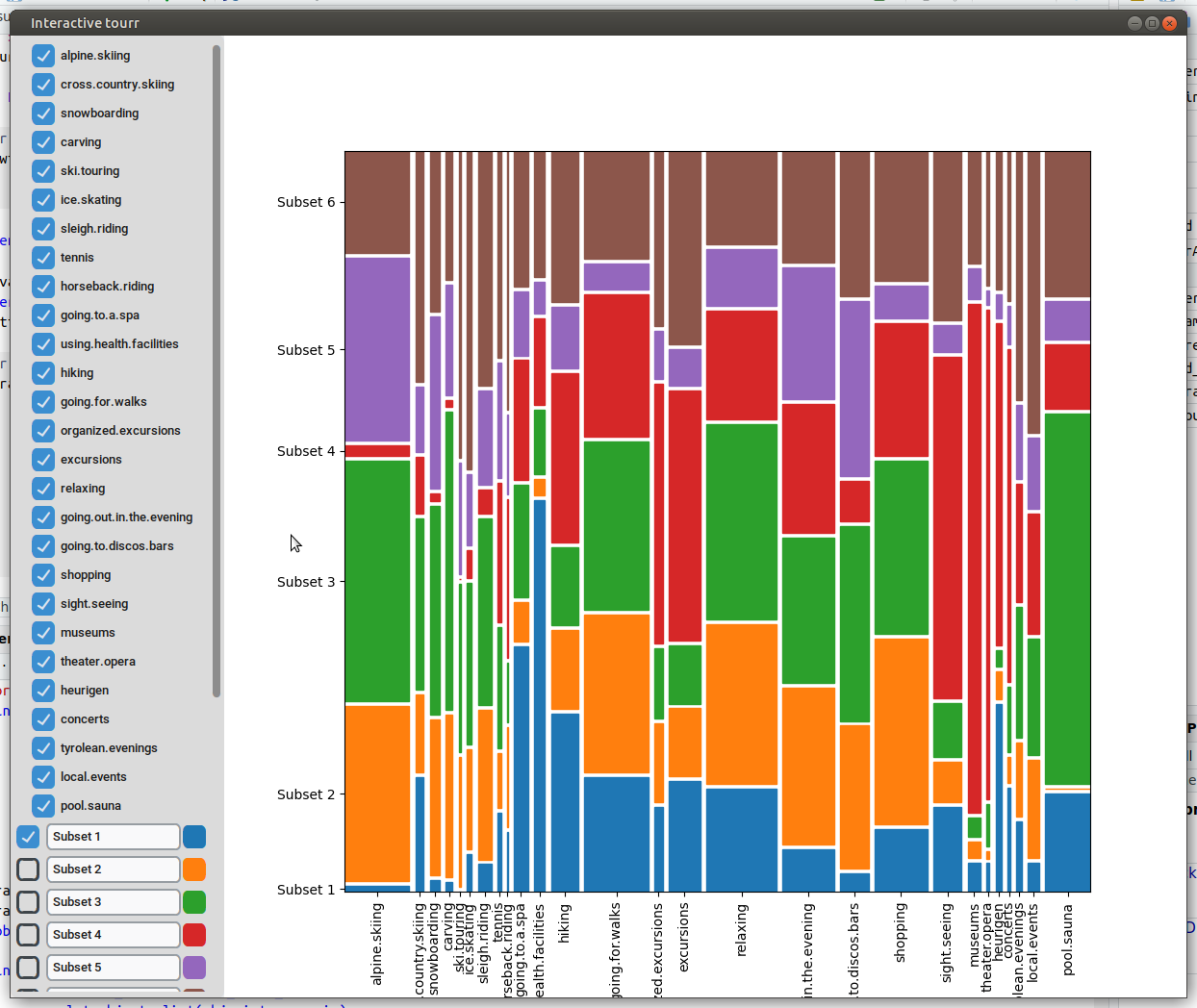
To display a mosaic plot, one has to provide whether the subgroups/clusters should be on the x or y axis, either with “subgroups_on_y” or “subgroups_on_x”.
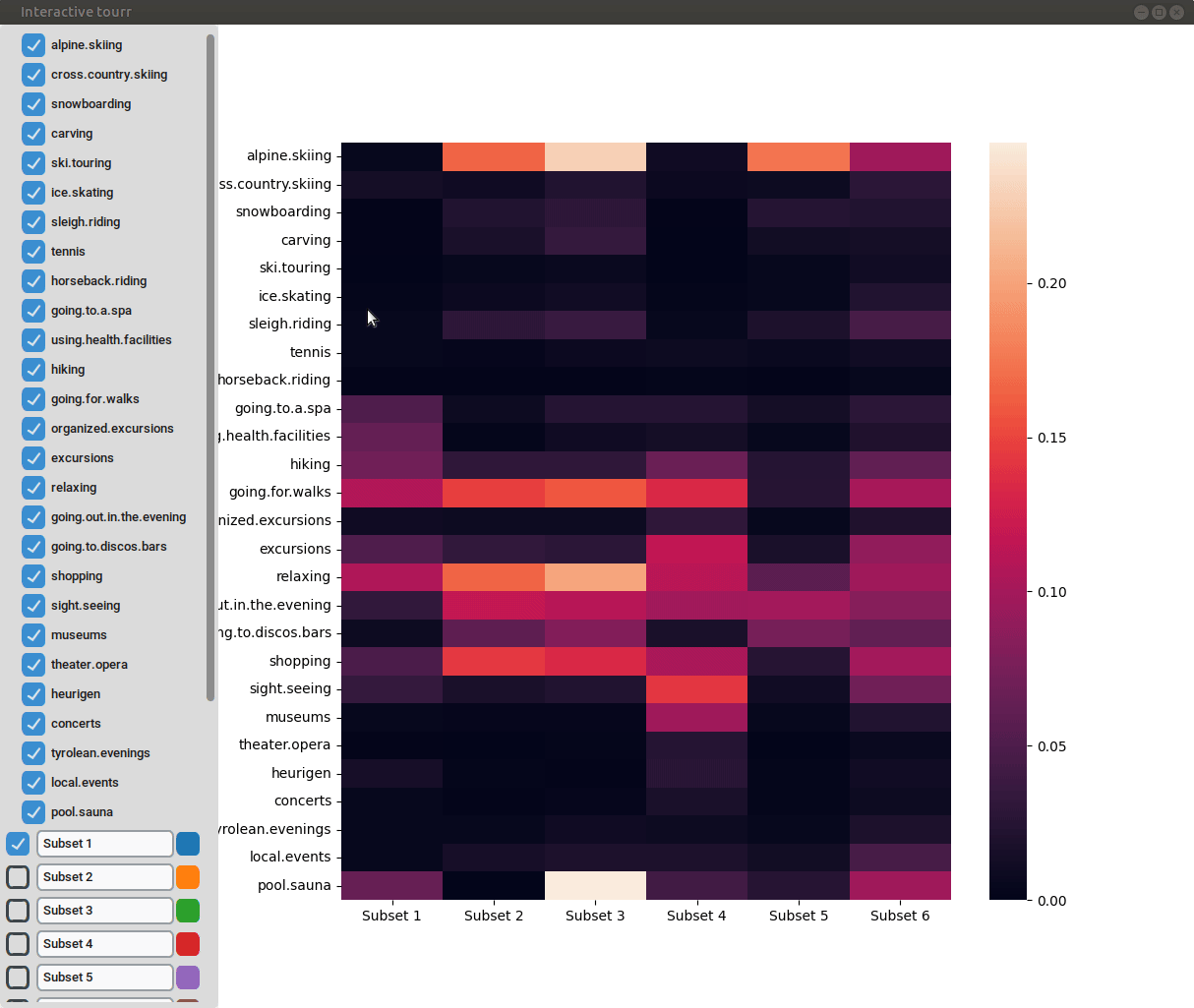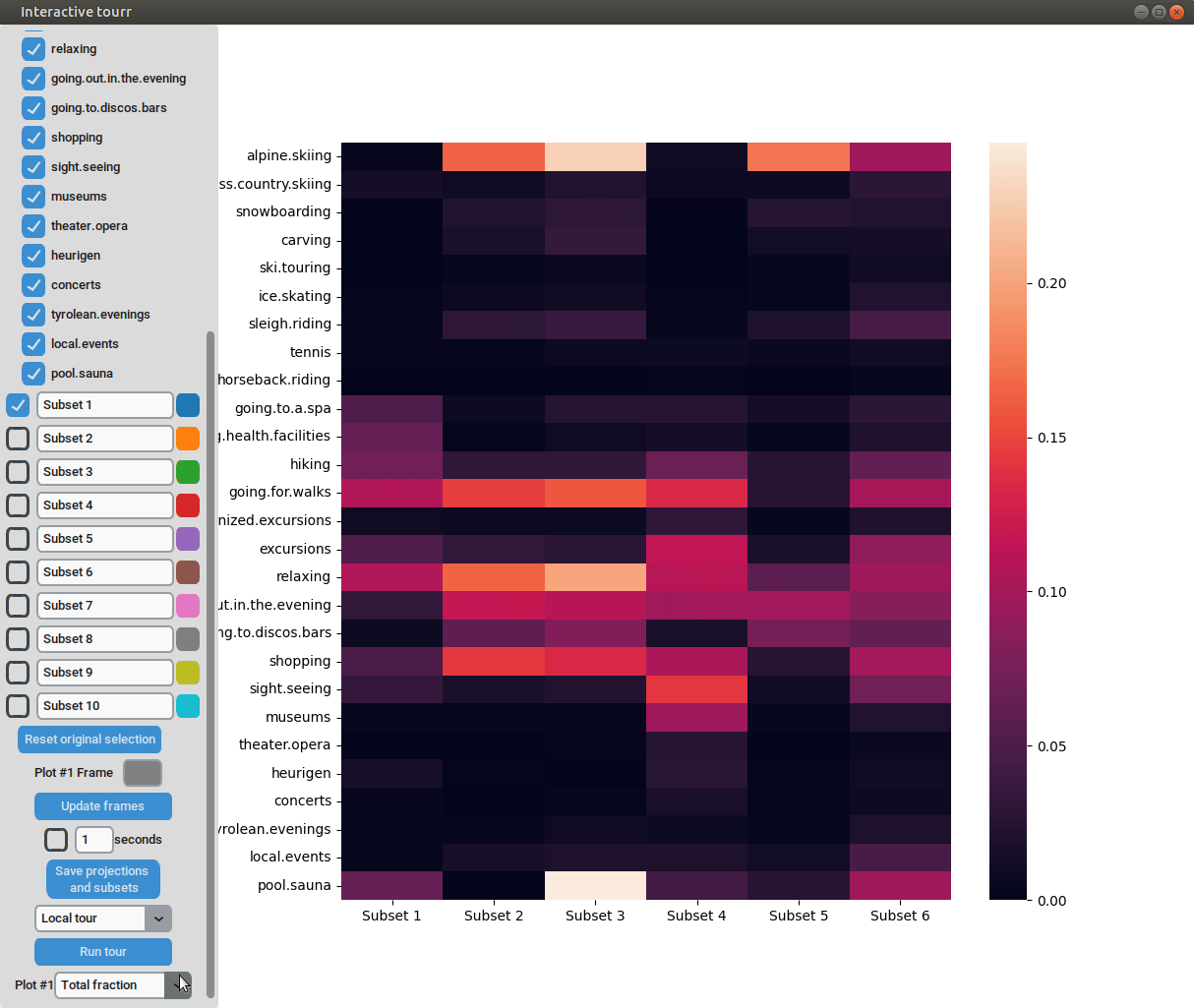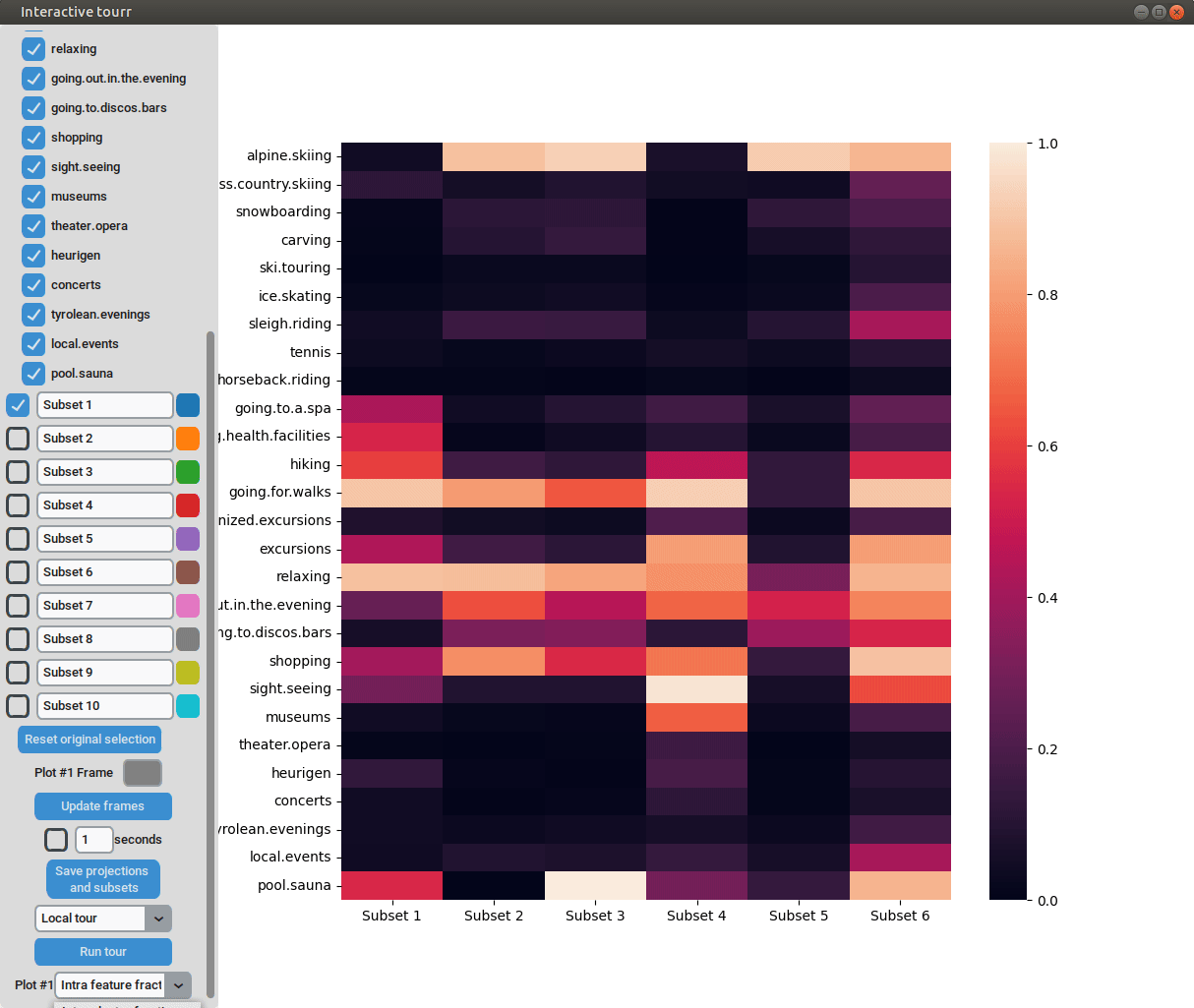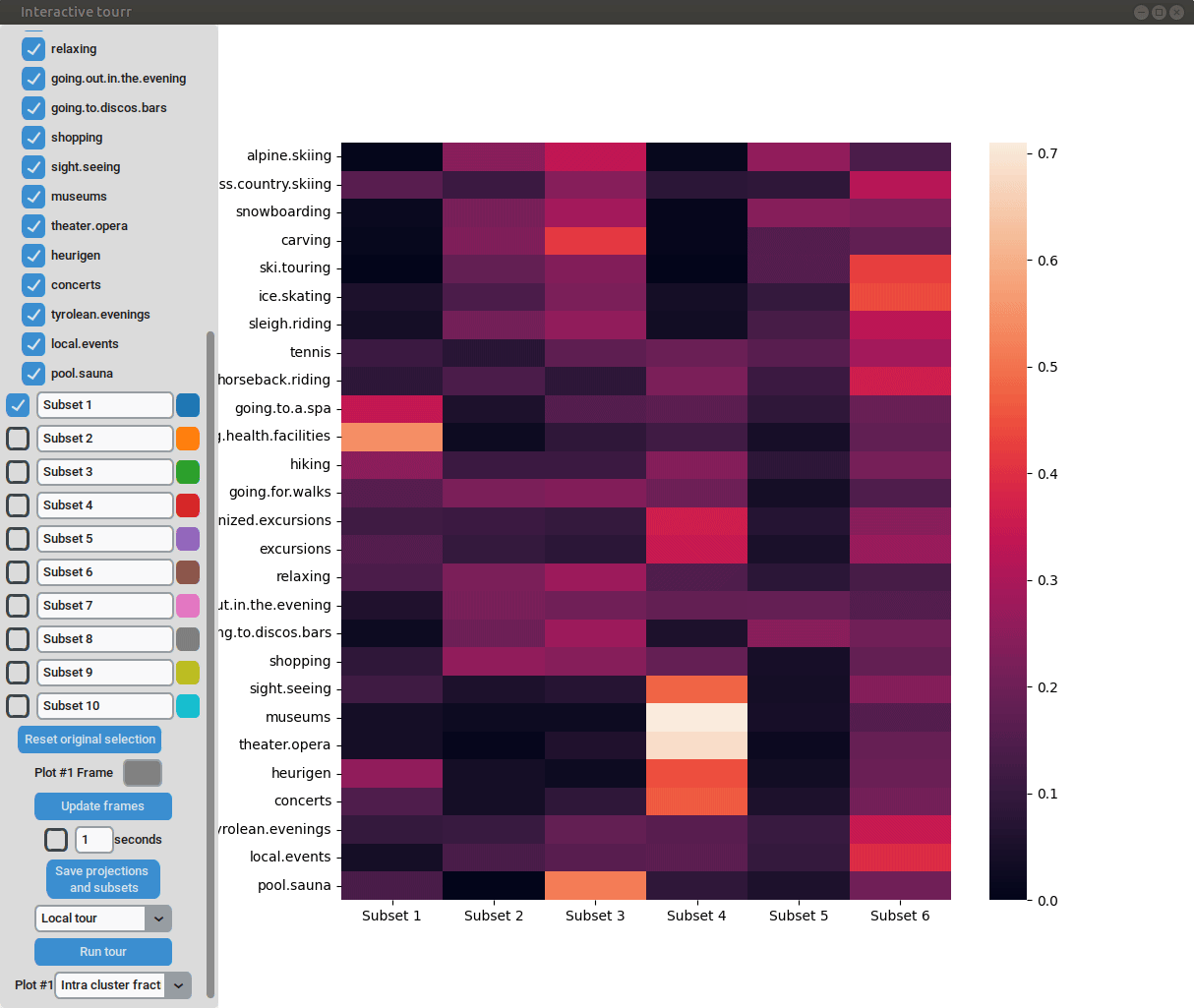
Heatmap
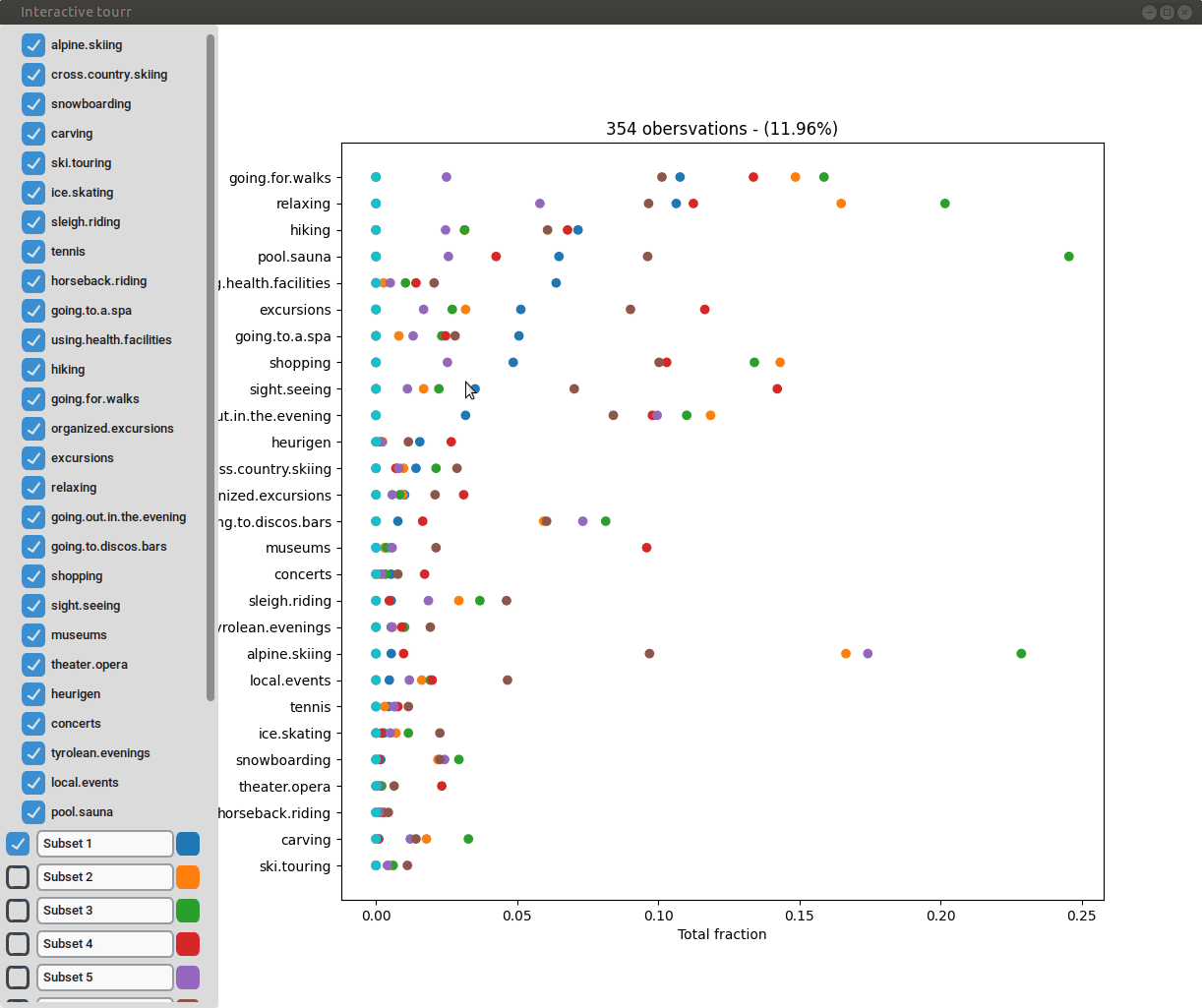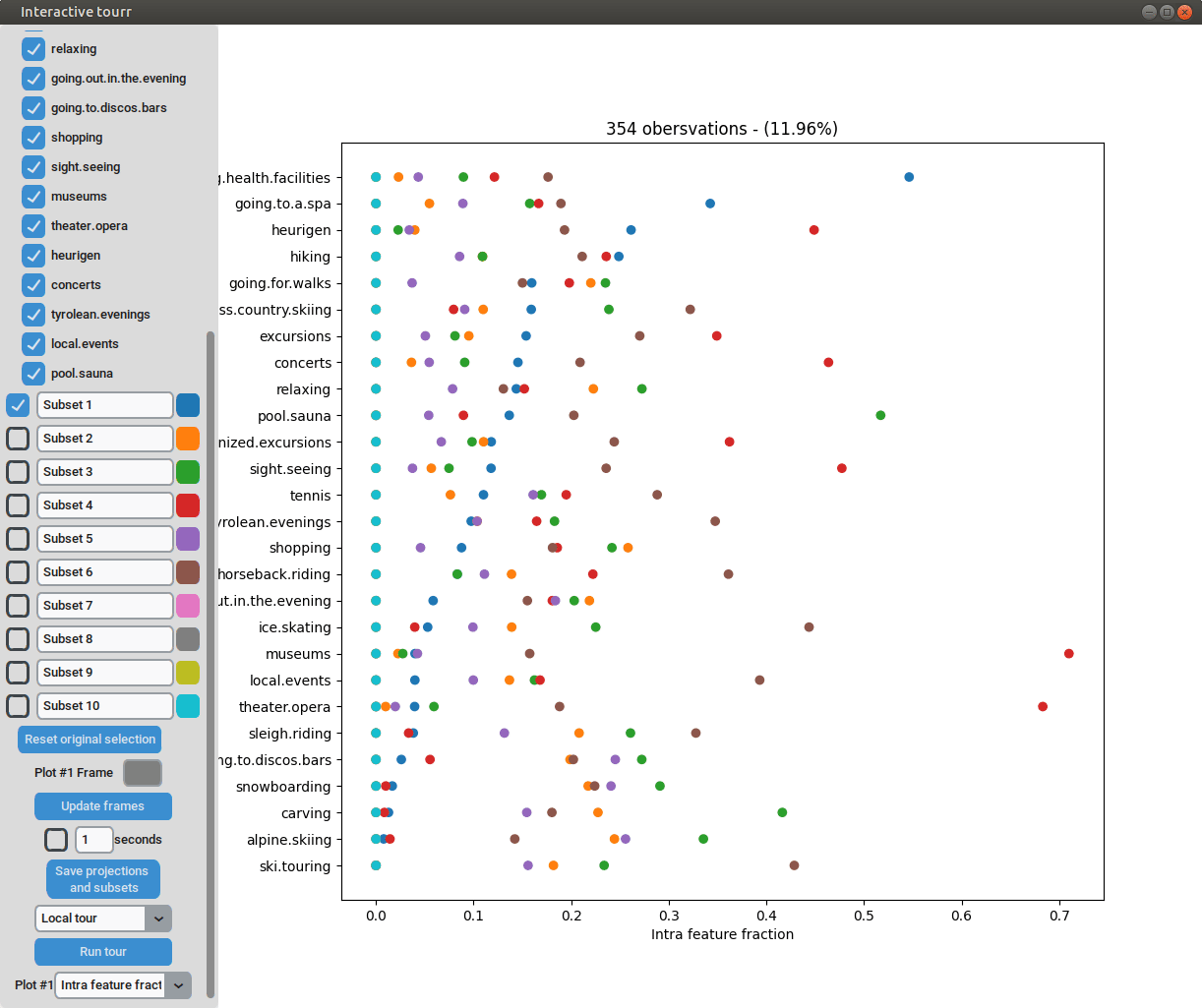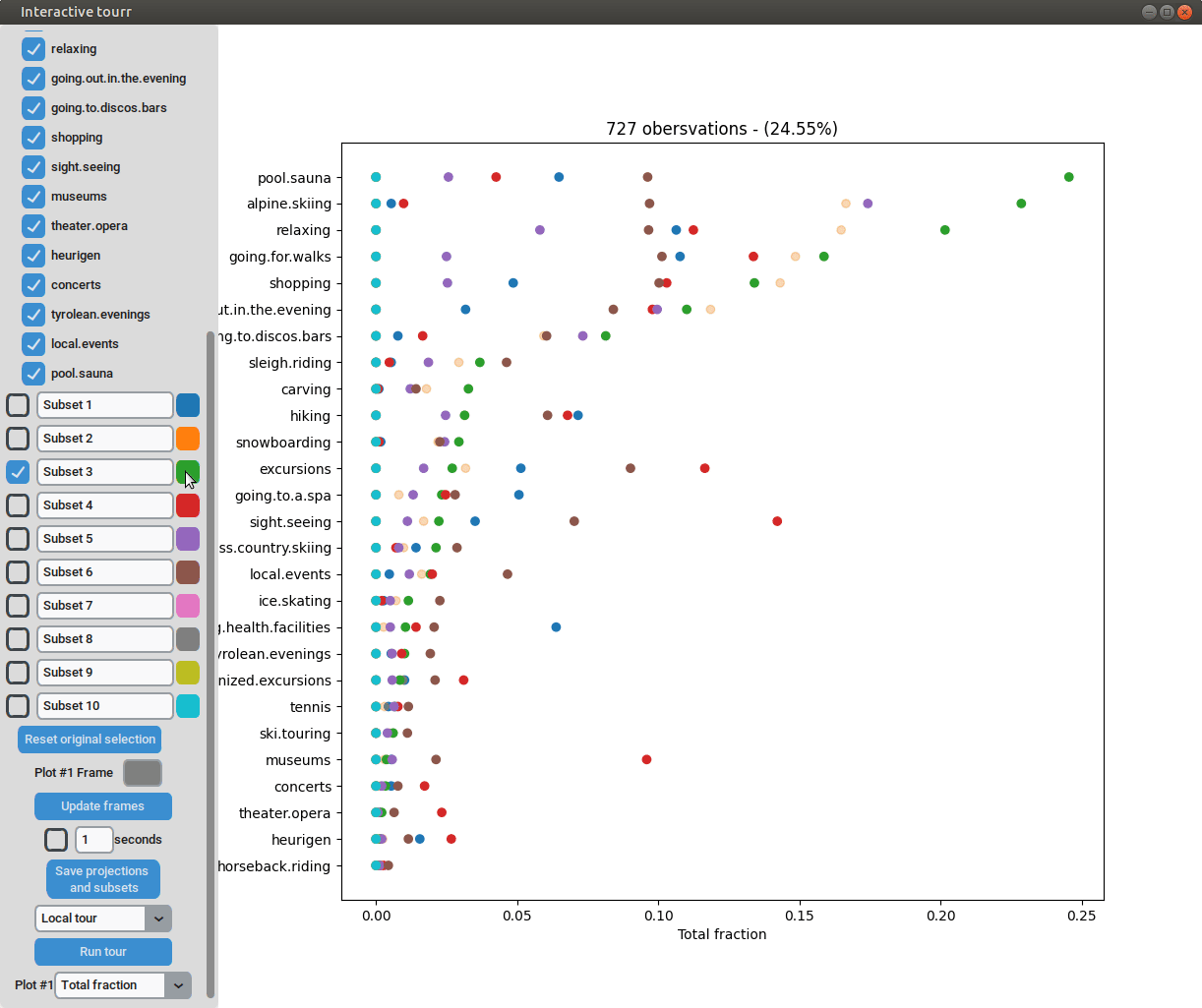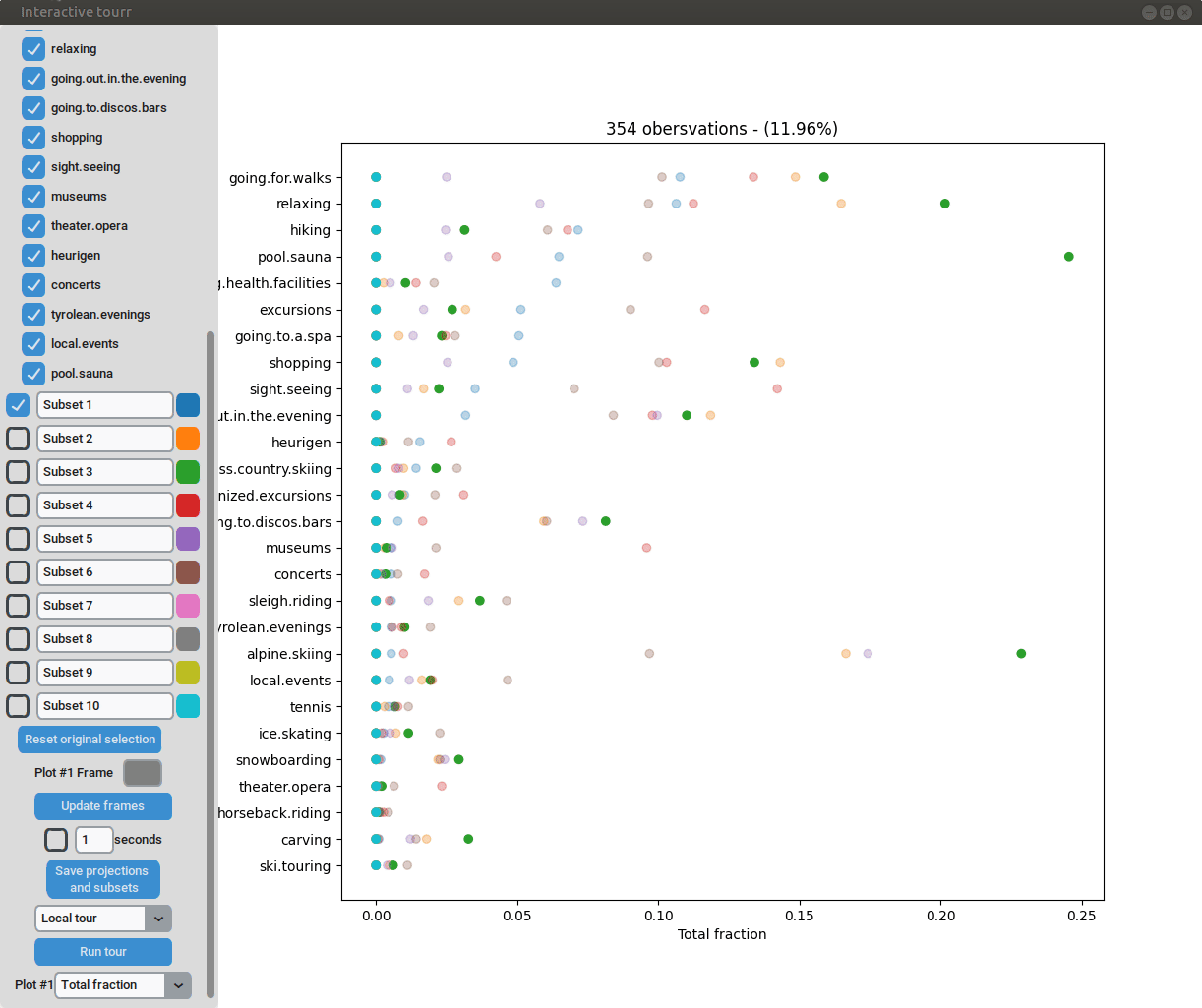
To display a heatmap, the metric to be calculated and plotted has to be selected. One can choose between “Total fraction”, “Intra cluster fraction” and “Intra feature fraction”.
Consider the matrix
where (number of clusters); (number of features) are a summary of each feature in each cluster. In case of binary data are the positive counts of the cluster/feature combination.
Then the total fraction is calculated by
the intra cluster fraction by and
the intra feature fraction by ,
where are the row and column totals.
Generating the displays
The various plot objects can the be displayed with the interactive_tour function.
# interactive_tour call of flea dataset. Insert plot objects of your liking.
if (interactive()){
interactive_tour(data=flea,
feature_names=feature_names_flea,
plot_objects=list(obj_flea_2d_tour),
half_range=half_range_flea,
n_plot_cols=2,
preselection=clusters_flea,
n_subsets=3,
preselection_names=flea_subspecies,
display_size=5)
}
# interactive_tour call of winterActiv dataset. Insert plot objects of your liking.
if (interactive()){
interactive_tour(data=winterActiv,
feature_names=feature_names_winter,
plot_objects=list(obj_winter_cat_clust),
half_range=3,
n_plot_cols=2,
preselection=clusters_winter,
n_subsets=10,
display_size=5)
}